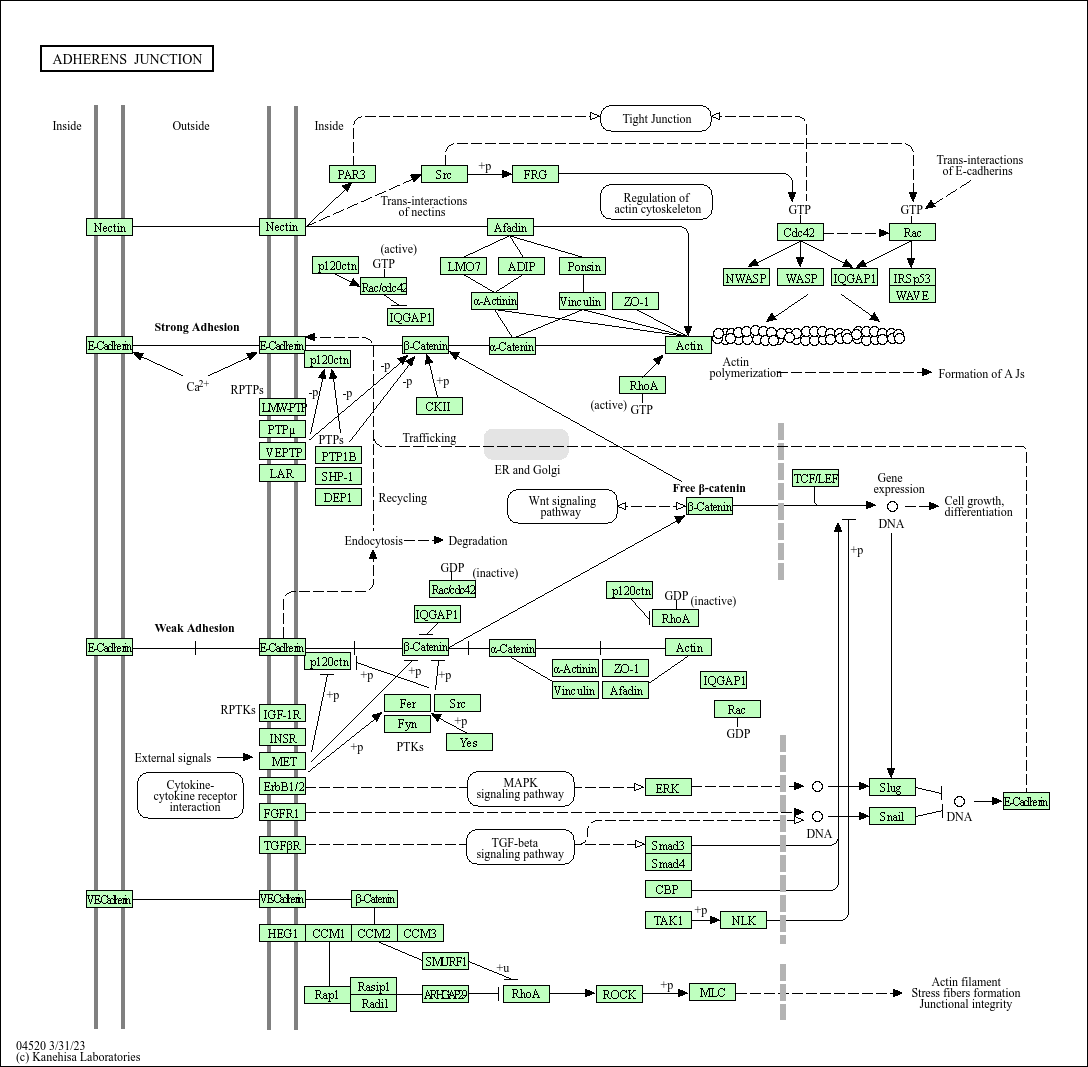
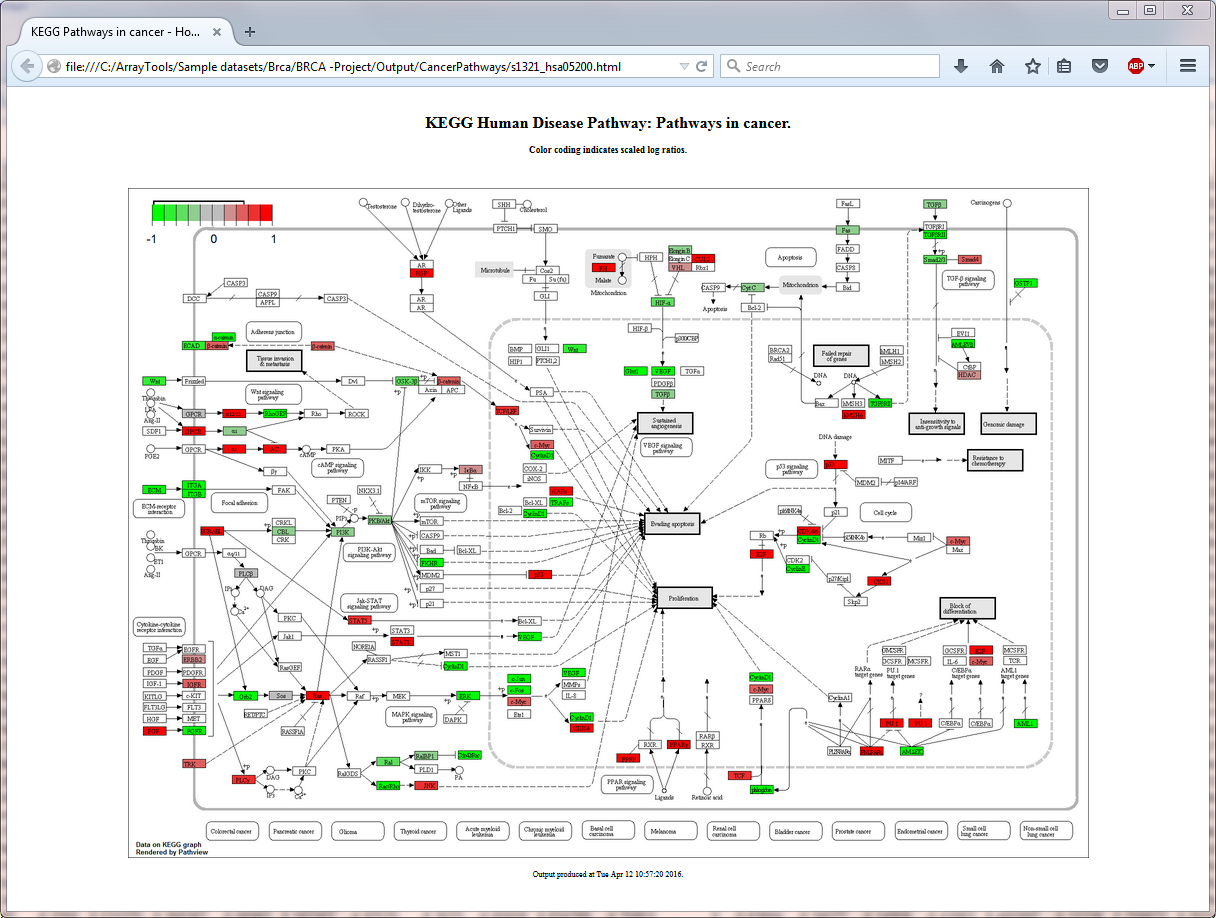
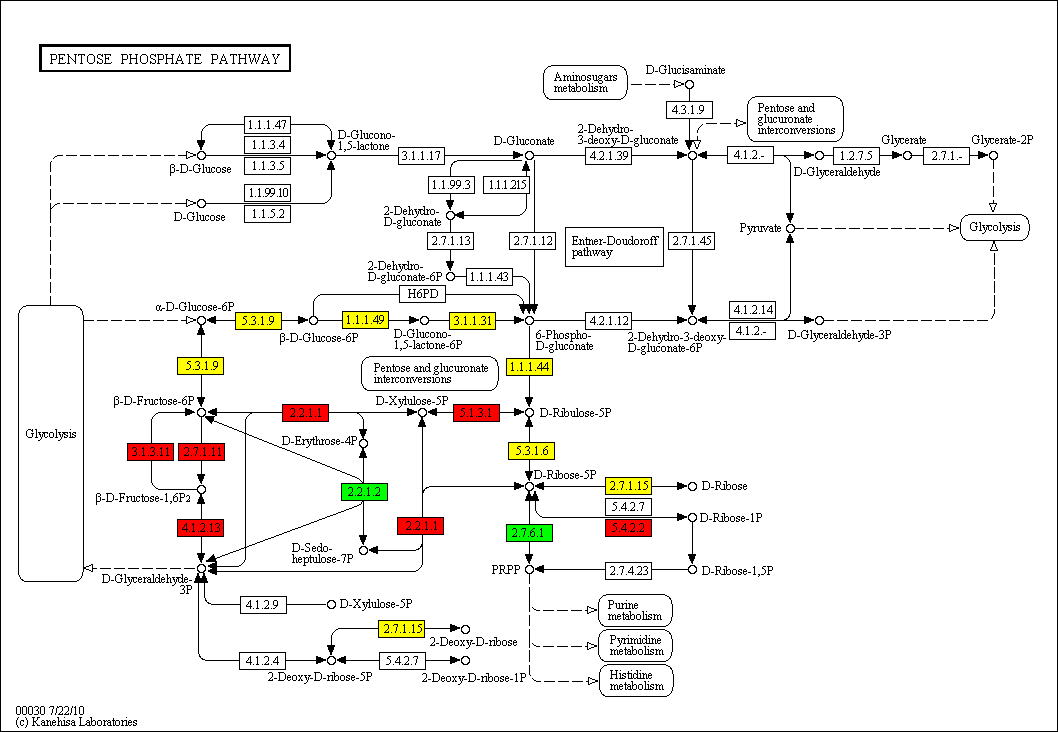
Kegg Pathway. KEGG (Kyoto Encyclopedia of Genes and Genomes) is a collection of databases dealing with genomes, biological pathways, diseases, drugs, and chemical substances. The KEGG pathway map is a moleculalr interaction/reaction network diagram represented in terms of the KEGG Orthology (KO) groups, so that experimental evidence in specific organisms can be.
At the DNA replication fork, a DNA helicase (DnaB or MCM complex) precedes the DNA synthetic machinery and unwinds the duplex.
If you have any questions, comments, or need information about CGAP, please contact.
In contrast to KEGG web, you can edit the network and map your data as you like. kegg-animate-pathway is a Python-based command-line utility which interfaces with the KEGG API to produce animated figures of metabolic pathways to reveal activities of its genes and/or compounds. How to use pathway database to understand disease? List of KEGG signaling pathways of human.